Projects
Gene Ontology Meta Annotator for Plants (GOMAP) is a high-throughput pipeline to annotate GO terms to plant protein sequences. The pipeline uses three different approaches to annotate GO terms to plant protein sequences, and compile the annotations together to generate an aggregated dataset. GOMAP uses Python code to run the component tools to generate the GO annotations, and R code to clean and aggregate the GO annotations from the component tools.
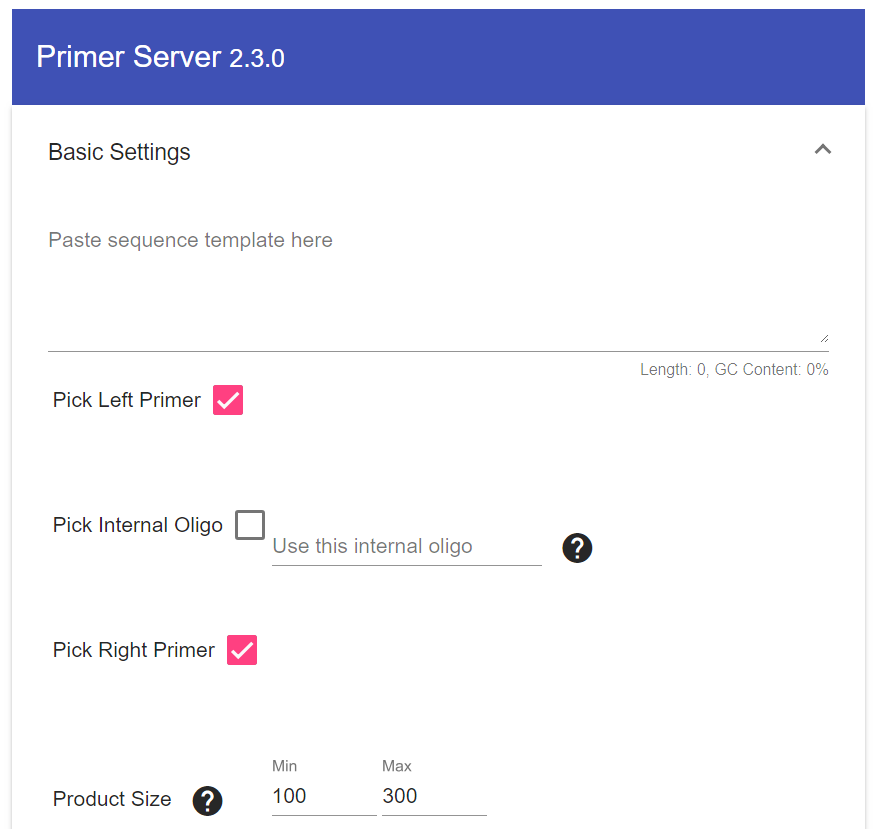
PrimerServer is an online tool to design specific primers for complex-genomes. Polymerase Chain Reaction (PCR) is a technique used to amplify specific DNA or RNA moecules for various tasks. Although many tools exist for designing PCR primers, not many tools are available to design specific primers against complex-genomes such as maize and animals. We implemented a easy to use web tool that uses Primer3 to design for a given input sequence and filters out non-specific primers by using BLAST to search against the genome.
maize-GAMER is a collaborative project to improve the status of gene functional annotation in maize. The project has three main areas of focus, namely design a pipeline for the functional annotation of maize genes, Use manually curated test data to evaluate the annotations and generate a best subset of annotations for use, and design a user friendly review system for the community to provide feedback and endorsements of the annotations
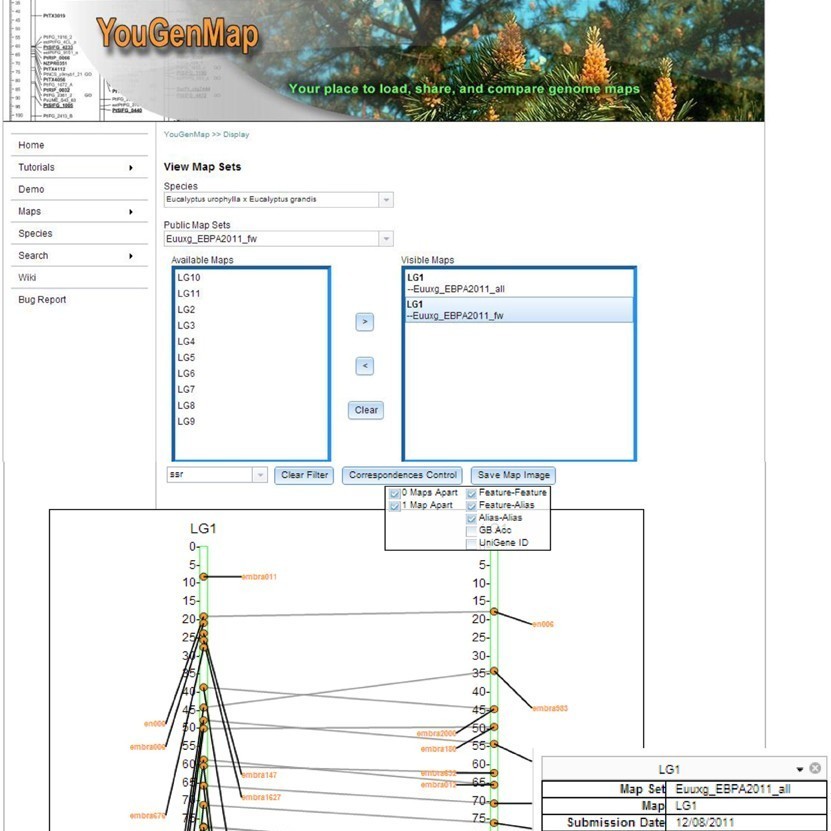
Comparative genetic maps are used in examination of genome organization, detection of conserved gene order, and exploration of marker order variations. YouGenMap is an open-source web tool that offers dynamic comparative mapping capability of users’ own genetic mapping between 2 or more map sets.
Website development for the Willard Sherman Turrell Herbarium at Miami University of Ohio. The herbarium has an extensive collection of materials from diverse number of plant species.
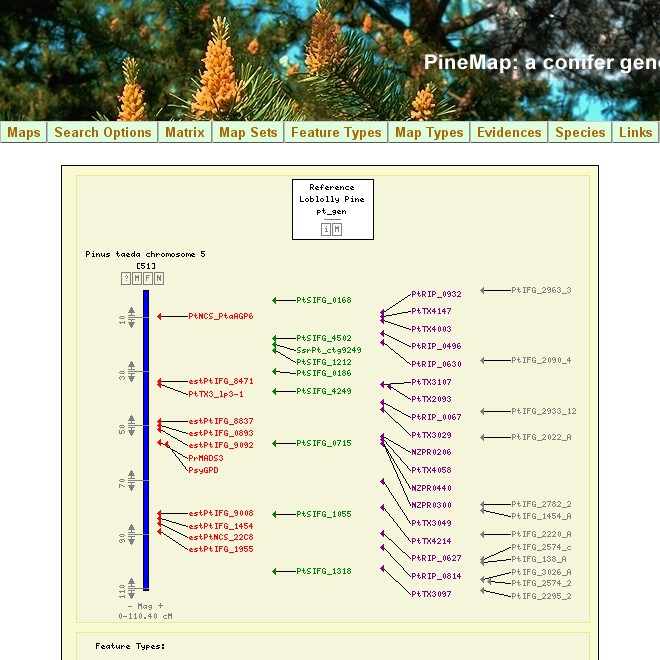
A collaborative project to implement CMap for Pinus taeda genetic maps. The generic mapping data generated for P. taeda, before the genome sequence was not available for public consumption. We used the open source genetic map visualization tool CMap from GMOD, and enabled public visualization of the dataset. This effort included heavy customization of the CMap code to be able to generate search functions.